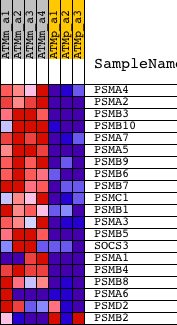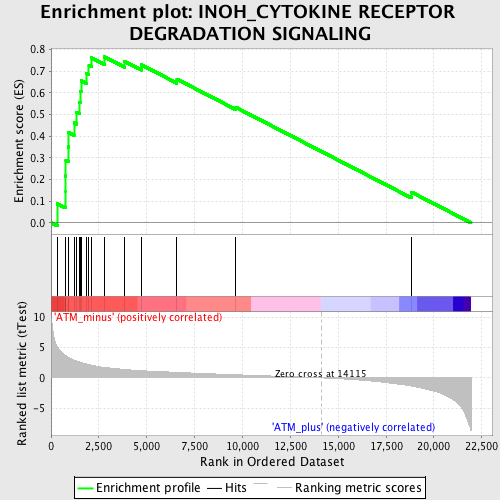
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | INOH_CYTOKINE RECEPTOR DEGRADATION SIGNALING |
| Enrichment Score (ES) | 0.766979 |
| Normalized Enrichment Score (NES) | 2.0413969 |
| Nominal p-value | 0.0 |
| FDR q-value | 7.1700674E-4 |
| FWER p-Value | 0.013 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMA4 | 1460339_at | 318 | 5.099 | 0.0883 | Yes | ||
| 2 | PSMA2 | 1448206_at | 744 | 3.668 | 0.1428 | Yes | ||
| 3 | PSMB3 | 1417052_at | 747 | 3.666 | 0.2166 | Yes | ||
| 4 | PSMB10 | 1448632_at | 773 | 3.615 | 0.2883 | Yes | ||
| 5 | PSMA7 | 1423567_a_at 1423568_at | 891 | 3.367 | 0.3509 | Yes | ||
| 6 | PSMA5 | 1424681_a_at 1434356_a_at | 919 | 3.308 | 0.4163 | Yes | ||
| 7 | PSMB9 | 1450696_at | 1201 | 2.903 | 0.4620 | Yes | ||
| 8 | PSMB6 | 1448822_at | 1328 | 2.765 | 0.5120 | Yes | ||
| 9 | PSMB7 | 1416240_at | 1485 | 2.609 | 0.5574 | Yes | ||
| 10 | PSMC1 | 1416005_at | 1516 | 2.575 | 0.6080 | Yes | ||
| 11 | PSMB1 | 1420052_x_at 1420053_at 1448166_a_at | 1566 | 2.522 | 0.6566 | Yes | ||
| 12 | PSMA3 | 1448442_a_at | 1853 | 2.256 | 0.6890 | Yes | ||
| 13 | PSMB5 | 1415676_a_at | 1978 | 2.164 | 0.7270 | Yes | ||
| 14 | SOCS3 | 1416576_at 1455899_x_at 1456212_x_at | 2112 | 2.071 | 0.7626 | Yes | ||
| 15 | PSMA1 | 1415695_at 1436769_at 1436770_x_at 1458292_at | 2780 | 1.726 | 0.7670 | Yes | ||
| 16 | PSMB4 | 1438984_x_at 1451205_at | 3848 | 1.368 | 0.7459 | No | ||
| 17 | PSMB8 | 1422962_a_at 1444619_x_at | 4723 | 1.187 | 0.7299 | No | ||
| 18 | PSMA6 | 1416506_at 1435316_at 1435317_x_at 1437144_x_at | 6574 | 0.902 | 0.6637 | No | ||
| 19 | PSMD2 | 1415831_at | 9652 | 0.509 | 0.5335 | No | ||
| 20 | PSMB2 | 1444377_at 1448262_at 1459641_at | 18820 | -1.334 | 0.1422 | No |